Image Analysis Support
Image Analysis is Hard.
Wako Automation is here to help
If you need assistance with any of the image analysis software below please use the “Contact Us” button at the top of this page and submit your request. A Wako Automation consultant will quickly get back to you.
Image Analysis Services
CellProfiler is cell image analysis software designed for biologists. The software was developed and is maintained by Dr. Anne Carpenter’s lab at the Broad Institute of Harvard and MIT.
CellProfiler is open source and free to download and use. Download here
In partnership with Dr. Carpenter’s lab, Wako Automation provides support to users of CellProfiler. Once you download CellProfiler try out example pipelines and tutorials. For additional assistance please visit the online forum.
Scientists from the CellProfiler community, Dr. Carpenter’s lab and Wako Automation continuously monitor the forum and provide answers to questions. If you need additional assistance, Wako Automation provides a number of services to help the imaging community.
CellProfiler training classes – A Fujifilm application scientist can provide a half day or more of training using a mix of example datasets as well as your own in-house data to get you on your way to building optimized CellProfiler pipelines/workflows designed to quantify images so you can advance your specific research projects.
CellProfiler pipeline design –
Some image analysis pipelines/workflows may be quite complex. In other instances your lab may be overwhelmed with imaging data and may need help to get you 90% of the way there. You might also need a pipeline designed from scratch. No matter your needs, Fujifilm can help.
This service is designed to help customers outsource CellProfiler pipeline/workflow design and simultaneously teach scientists how to become CellProfiler experts. Scientists upload a few sample images for a specific project (positive control, negative control, and sample images), along with an explanation of what needs to be quantified.
From these images, our application scientists will develop a CellProfiler pipeline/workflow to get you the results for your experiment. This pipeline is sent back to our customer who can then work with our application scientist to finalize an optimized pipeline. This process not only gets you a jump start on one of the most difficult aspects of running an imaging experiment, but also helps train you to become an CellProfiler expert.
Fiji/ImageJ, KNIME, QuPath, Ilastik, custom Python image analysis scripts – In addition to working with CellProfiler, Fujifilm Wako Automation can provide training and analysis assistance for Fiji/ImageJ, KNIME, Ilastik and QuPath. If your application requires a fully custom solution we can provide custom Python scripts to get you the results you are looking for.
Below are a number of free CellProfiler pipeline/workflow tutorials, created by our applications scientists, to assist in a broad range of image analysis challenges. Download the tutorials and see how our application scientists use CellProfiler to solve these challenges. Check back often as we add more pipeline/workflow tutorials.
Getting Started with CellProfiler
A basic guide for scientists who are using CellProfiler for the first time. Please download and review this guide and then download and follow the tutorials. If you already have CellProfiler experience, skip this guide and go right to the tutorials.
Download Tutorial »
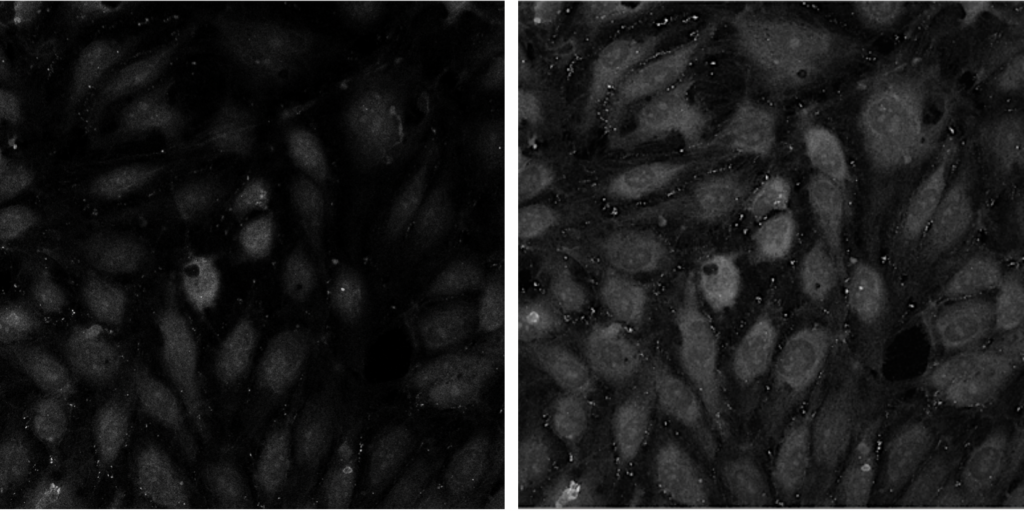
Segment Fibroblasts
Aim: To segment spindle shaped fibroblasts
This pipeline illustrates techniques used to segment spindle shaped fibroblasts from a three channel image using basic CellProfiler modules that enable image thresholding adjustments and smoothing parameters. In addition, colocalization of two of the channels from the segmented objects is reported.
Image: Multichannel image (DAPI, Actin, Myosin)
Download Tutorial »
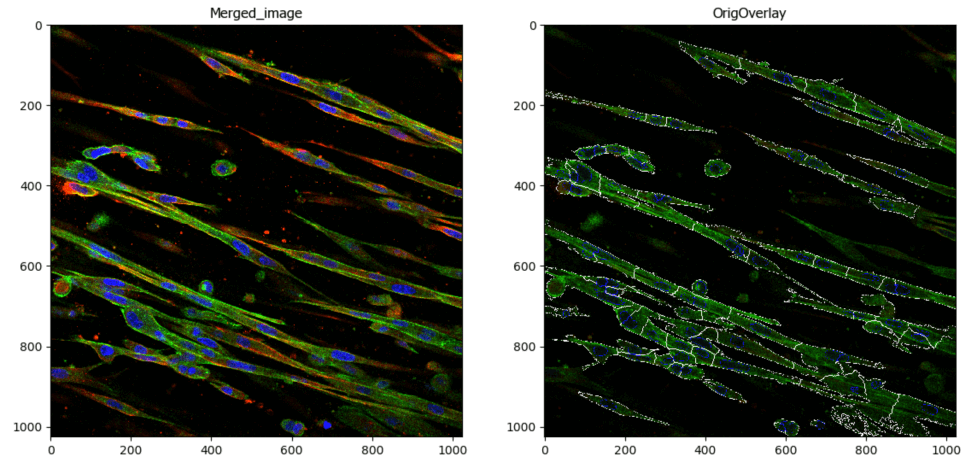
Segment Microglia
Aim: To segment microglia
This pipeline illustrates the segmentation of complete microglia with dendrites using CellProfiler’s™ EnhanceSuppress features.
Image: 2 channel image (DAPI & GFP)
Download Tutorial »
3D Segmentation of Tumor Cells
Aim: to segment cells in 3D
This pipeline illustrates how CellProfiler can be used to perform 3D analysis on confocal images of cells and nuclei.
Image: two channel confocal image
Download Tutorial »
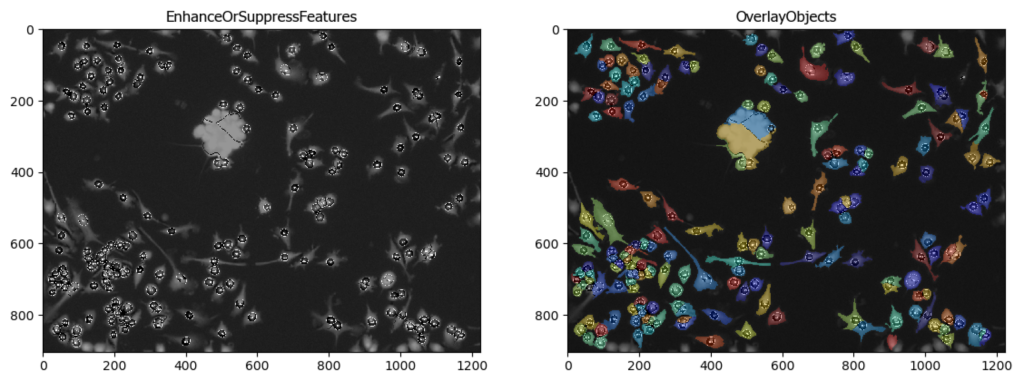
Segmentation of Cells in Tissue with High Background
Aim: Segment cells within a tissue image with high background.
This pipeline utilizes CellProfiler’s EnhanceOrSuppressFeatures module to suppress high background before segmentation.
Image: A single tissue image.
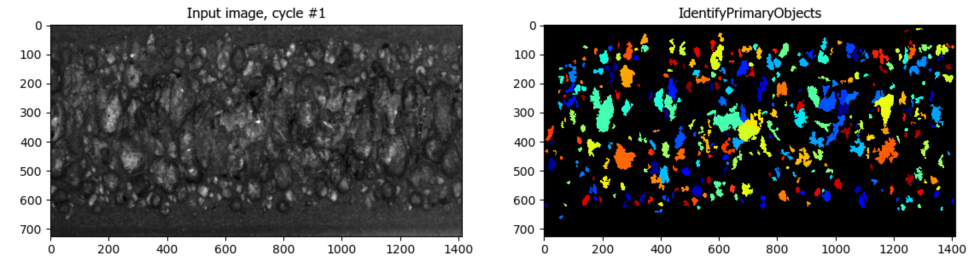
Download Tutorial »
Using the MaskObjects Function With A Manually Drawn or Imported Region Of Interest
Aim: Identify objects within a region of interest that is either manually drawn within CellProfiler or has been pre-saved as a mask.
Images: One multi-colored neuronal image and a binary image of a predefined mask representing the Region Of Interest (ROI).
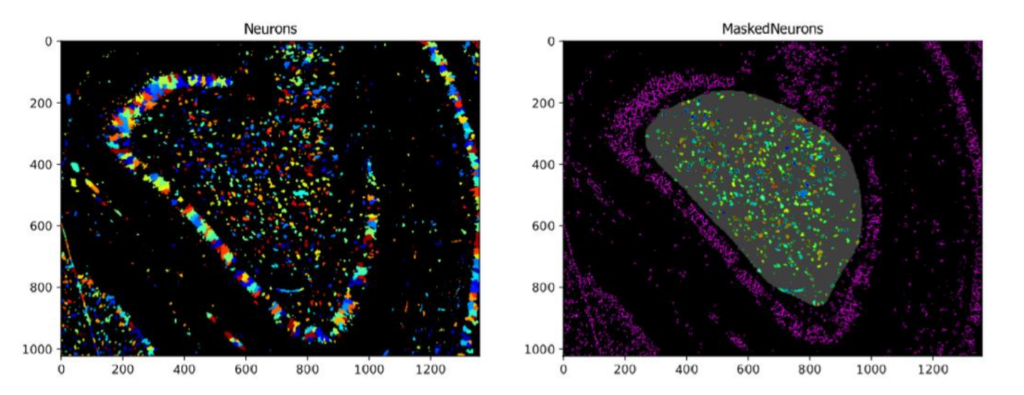
Download Tutorial »
Seed counting (Agriculture Application)
Aim: To identify, segment, and count corn kernels from the image of an ear of a corn.
This pipeline illustrates CellProfiler’sTM ability to identify the local maxima from a standard photograph. By identifying local maxima corn kernels can be recognized and counted.
Image: Single color image of an ear of corn.

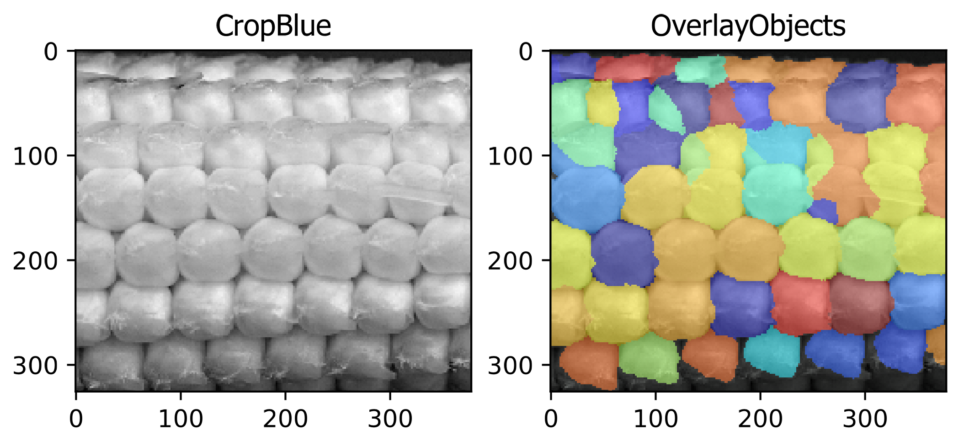
Download Tutorial »
Segmentation and Classification of Individual Oocytes Based on Size
Aim: Segment individual oocytes and classify them by size.
This pipeline illustrates how to create a mask of the region of interest (in this case the tissue), to isolate a colony of oocytes, identify and segment individual oocytes, and then filter them based on size.
Image: Single color image
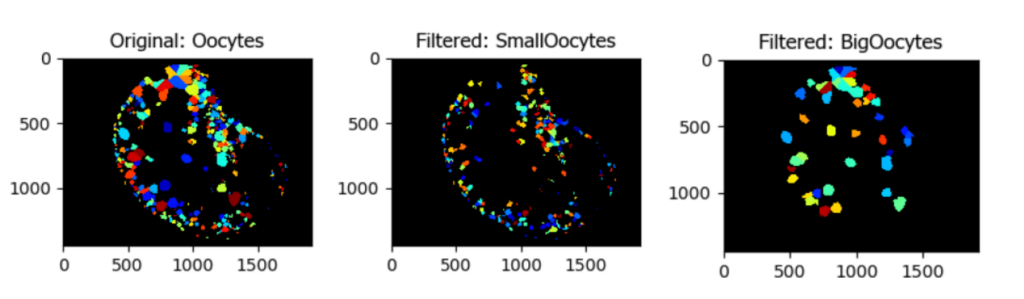
Download Tutorial »
Segmenting a Viral Plaque from a Brightfield Image
This example pipeline shows the segmentation of a large region, in this case a viral plaque, from a brightfield image.
Image: A Single Differential Interference Contrast (DIC) Image
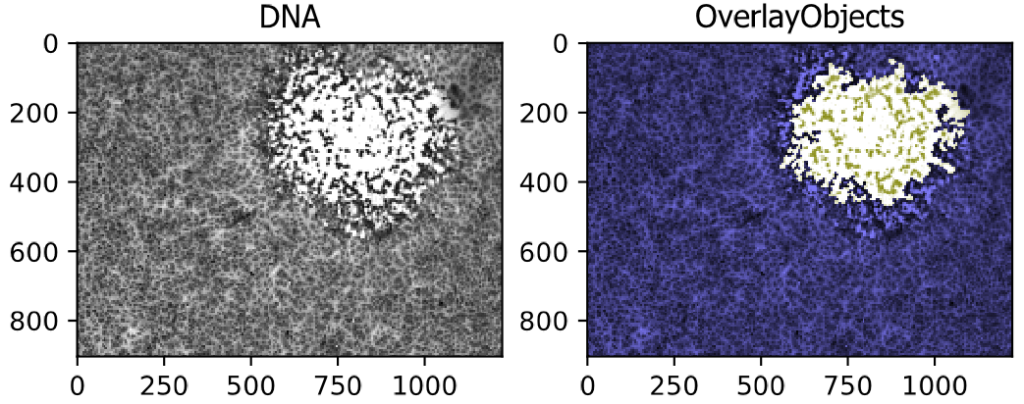
Download Tutorial »
Saving each Segmented Cell as an Individual Mask
Aim: Demonstrate CellProfiler’s unique ability to save each segmented cell as an individual mask. These saved masks can be used later within CellProfiler or in other image analysis applications.
Image: Single image with nuclei and cell signal
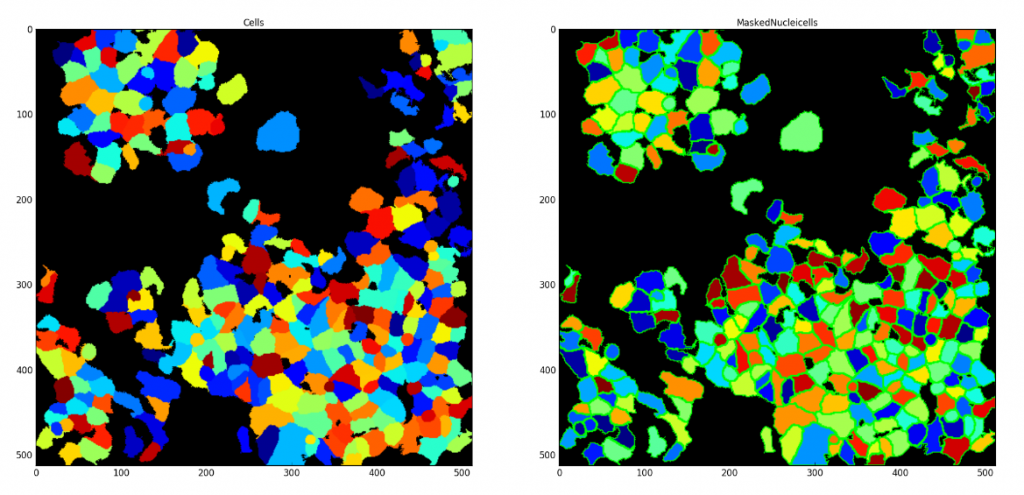
Download Tutorial »
Using the Groups Module to Analyze Data by Well
Aim: Utilize the Metadata, NamesAndTypes, and Groups modules to create Maximum Image Projections (MIPs) of Z stack images for three channels and two wells of data.